publications
2026
- bioRxiv
 Learning a PRECISE language for small-molecule bindingMert Erden*, Xuting Zhang*, Kapil Devkota*, Rohit Singh†, and Lenore Cowen†bioRxiv, 2026
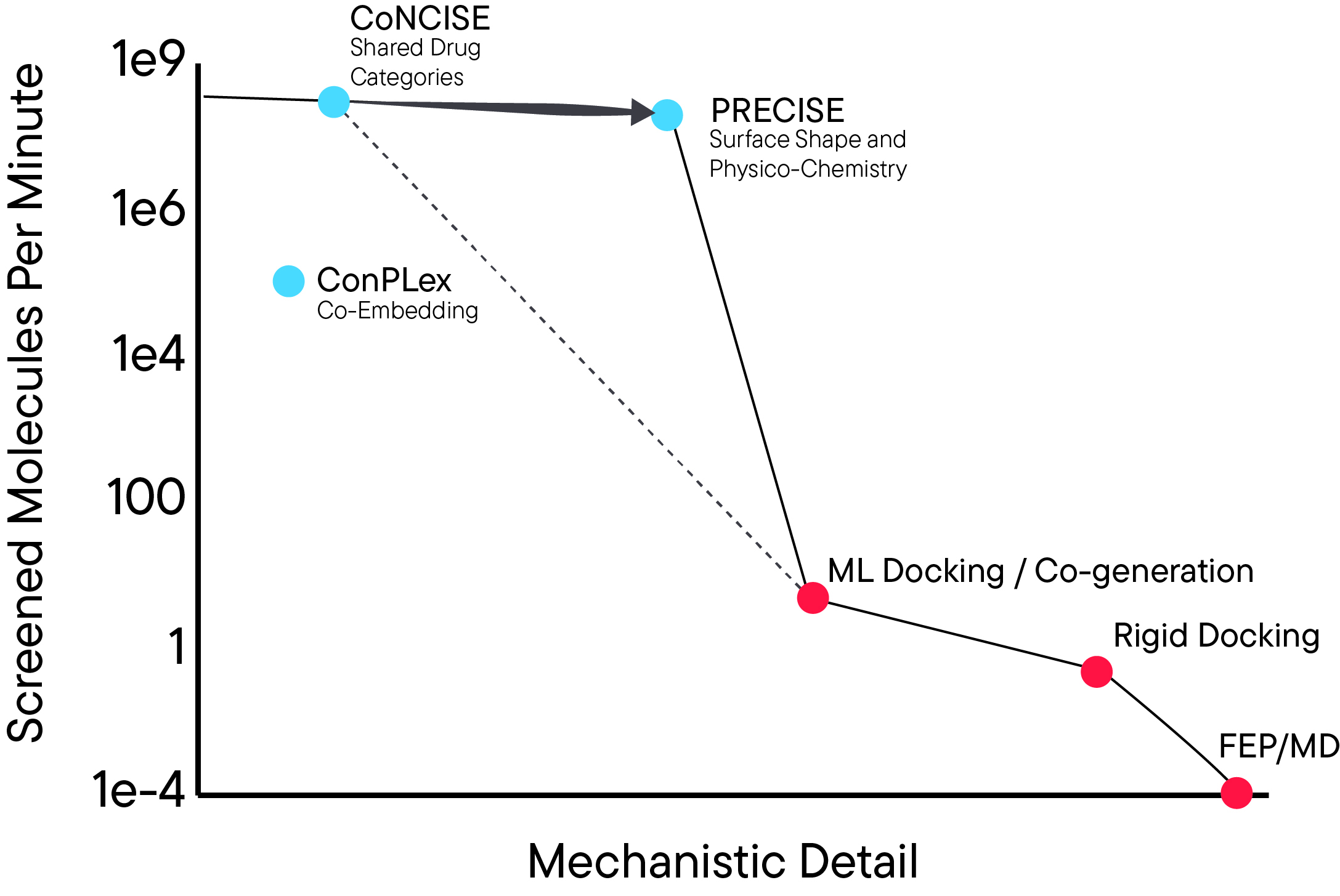
Learning a PRECISE language for small-molecule bindingMert Erden*, Xuting Zhang*, Kapil Devkota*, Rohit Singh†, and Lenore Cowen†bioRxiv, 2026Virtual screening of billion-scale compound libraries has become feasible through machine learning approaches. In particular, CoNCISE (RECOMB 2025) introduced drug quantization via codebooks, achieving highly scalable and accurate binary predictions. However, drug discovery requires understanding not just whether molecules bind, but where they bind and how to target specific sites. Here, we present PRECISE which leverages CoNCISE’s quantized small-molecule representations while operating on the target’s 3D structure as its input. The key innovation of PRECISE is reimagining drug-target interaction as compatibility between quantized drug embeddings and a latent representation of the target’s surface mesh, enriched with electrostatic and geometric features. PRECISE designs a novel surface representation, interpreted through a geometric deep learning architecture, enabling it to identify binding sites more accurately than state-of-the-art methods (DiffDock-L, Chai, and Boltz-2) while the codebook ensures billion-scale screening capability. Our formulation unlocks zero-shot generalization to complex targets such as metalloproteins and multi-chain complexes. To enable efficient integration with downstream docking workflows, we introduce PRECISE-MCTS, which combines fast PRECISE-based screening with selective Vina docking through an iterative Monte Carlo Tree Search approach. By providing both mechanistic understanding and massive scalability, PRECISE delivers capabilities that were previously mutually exclusive in virtual screening.Competing Interest StatementThe authors have declared no competing interest.
@article{Erden2026.01.04.697581, author = {Erden, Mert and Zhang, Xuting and Devkota, Kapil and Singh, Rohit and Cowen, Lenore}, title = {Learning a PRECISE language for small-molecule binding}, elocation-id = {2026.01.04.697581}, year = {2026}, doi = {10.64898/2026.01.04.697581}, publisher = {Cold Spring Harbor Laboratory}, url = {https://www.biorxiv.org/content/early/2026/01/05/2026.01.04.697581}, eprint = {https://www.biorxiv.org/content/early/2026/01/05/2026.01.04.697581.full.pdf}, journal = {bioRxiv}, }
2025
- Nat. Biotech.
 A trimodal protein language model enables advanced protein searchesJin Su†, Yan He†, Shiyang You†, Shiyu Jiang, Xibin Zhou, Xuting Zhang, Yuxuan Wang, Xining Su, Igor Tolstoy, Xing Chang, and othersNature Biotechnology, 2025
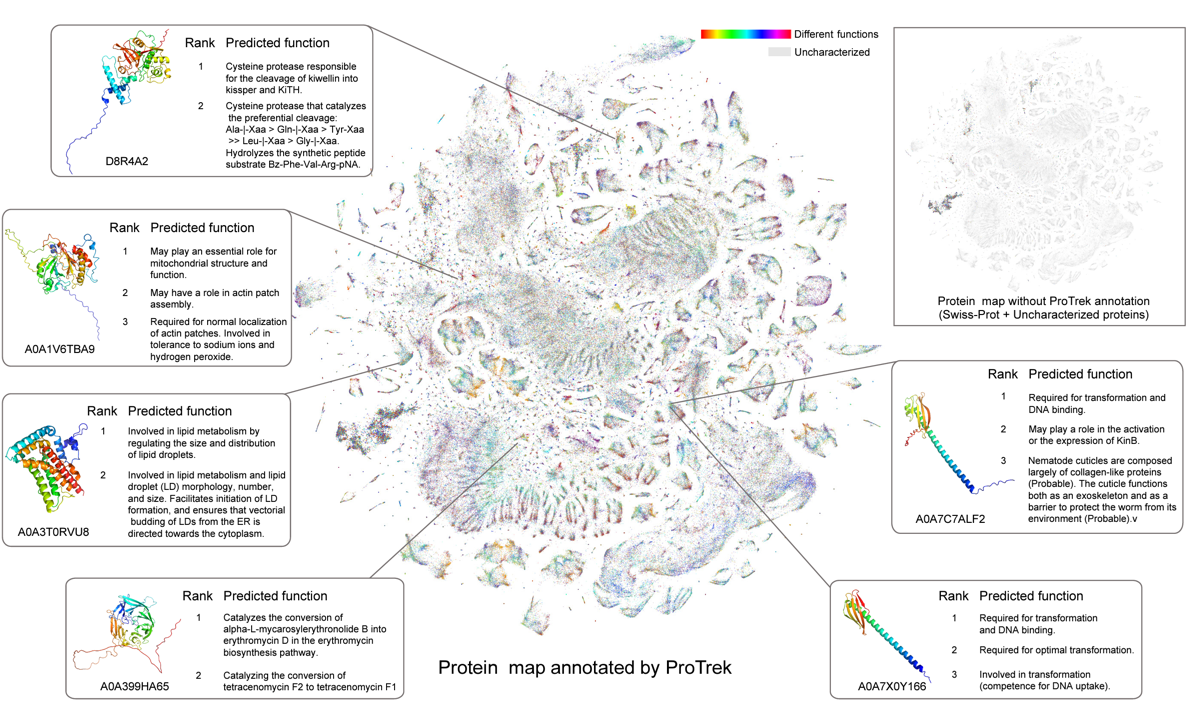
A trimodal protein language model enables advanced protein searchesJin Su†, Yan He†, Shiyang You†, Shiyu Jiang, Xibin Zhou, Xuting Zhang, Yuxuan Wang, Xining Su, Igor Tolstoy, Xing Chang, and othersNature Biotechnology, 2025@article{su2025trimodal, title = {A trimodal protein language model enables advanced protein searches}, author = {Su, Jin and He, Yan and You, Shiyang and Jiang, Shiyu and Zhou, Xibin and Zhang, Xuting and Wang, Yuxuan and Su, Xining and Tolstoy, Igor and Chang, Xing and others}, journal = {Nature Biotechnology}, pages = {1--7}, year = {2025}, publisher = {Nature Publishing Group US New York}, } - Nat. Biotech.
 Democratizing protein language model training, sharing and collaborationJin Su, Zhikai Li, Tianli Tao, Chenchen Han, Yan He, Fengyuan Dai, Qingyan Yuan, Yuan Gao, Tong Si, Xuting Zhang, and othersNature Biotechnology, 2025
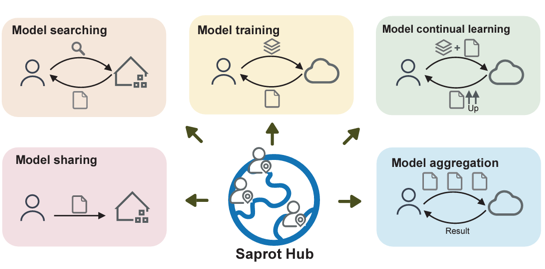
Democratizing protein language model training, sharing and collaborationJin Su, Zhikai Li, Tianli Tao, Chenchen Han, Yan He, Fengyuan Dai, Qingyan Yuan, Yuan Gao, Tong Si, Xuting Zhang, and othersNature Biotechnology, 2025@article{su2025democratizing, title = {Democratizing protein language model training, sharing and collaboration}, author = {Su, Jin and Li, Zhikai and Tao, Tianli and Han, Chenchen and He, Yan and Dai, Fengyuan and Yuan, Qingyan and Gao, Yuan and Si, Tong and Zhang, Xuting and others}, journal = {Nature Biotechnology}, pages = {1--7}, year = {2025}, publisher = {Nature Publishing Group US New York}, } - bioRxiv
 STARNet enables spatially resolved inference of gene regulatory networks from spatial multi-omics dataLei Hu†, Shichen Zhang†, Xuting Zhang†, Yihai Luo†, Haoteng Gu, Peng Liu, Sheng Mao, Li Chen, Yuhao Xia, Minghao Yang, and othersbioRxiv, 2025
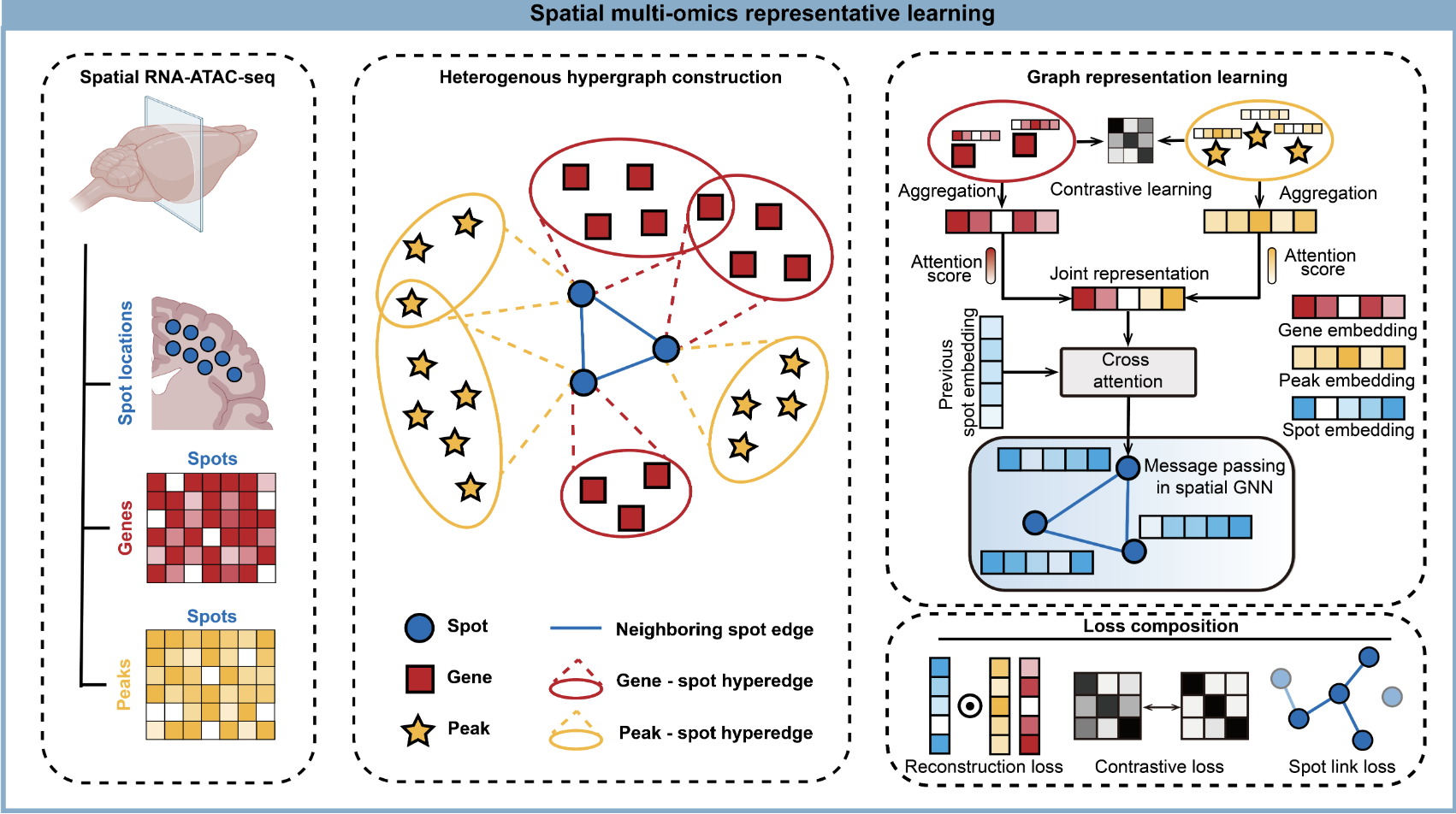
STARNet enables spatially resolved inference of gene regulatory networks from spatial multi-omics dataLei Hu†, Shichen Zhang†, Xuting Zhang†, Yihai Luo†, Haoteng Gu, Peng Liu, Sheng Mao, Li Chen, Yuhao Xia, Minghao Yang, and othersbioRxiv, 2025Biological tissues are composed of distinct microenvironments that spatially orchestrate gene expression and cell identity. However, the regulatory principles governing domain-specific cellular functions remain poorly understood due to the lack of effective methods for mapping gene regulatory networks (GRNs) in situ. To address this gap, we introduce STARNet, a representation learning approach that leverages heterogeneous hypergraph modeling of spatial transcriptomic and epigenomic data to resolve tissue-domain-specific regulatory interactions. By integrating graph neural networks with contrastive learning in a self-supervised framework, STARNet learns unified embeddings that preserve both multi-modal molecular features and anatomical spatial context, enabling accurate and domain-resolved GRN reconstruction within complex tissues. Benchmarking on both simulated and real datasets demonstrates that STARNet achieves state-of-the-art performance. We further demonstrate its broad applicability across diverse biological contexts, including neural development, genetic disease risk, and drug-induced developmental toxicity. In the mouse brain, it delineates region-specific regulatory networks and reconstructs spatiotemporal programs underlying neural stem cell differentiation. In human genetics, it provides a mechanistic link between genotypes and phenotypes by showing how genome-wide association study (GWAS) variants for complex diseases perturb hippocampus-specific GRNs. In developmental toxicology, STARNet reveals that drug-induced disruptions of GRNs in defined embryonic regions underlie tissue-specific vulnerability. Collectively, STARNet offers a powerful and versatile framework for resolving the spatial regulatory logic of complex tissues, providing multi-angle insights into tissue patterning, development, and disease mechanisms.
@article{hu2025starnet, title = {STARNet enables spatially resolved inference of gene regulatory networks from spatial multi-omics data}, author = {Hu, Lei and Zhang, Shichen and Zhang, Xuting and Luo, Yihai and Gu, Haoteng and Liu, Peng and Mao, Sheng and Chen, Li and Xia, Yuhao and Yang, Minghao and others}, journal = {bioRxiv}, pages = {2025--08}, year = {2025}, publisher = {Cold Spring Harbor Laboratory}, }